14.1.4.5. Fick’s Law Data Model Scripting Example¶
MCell and CellBlender can be used to demonstrate various aspects of Fick’s 1st and 2nd laws. The Advanced Models section contains a tutorial on constructing such a model in CellBlender by hand. The following Data Model Script constructs an entire Fick’s demonstration model in Python.
As shown in the animation below, the script constructs a long semi-transparent blue box which will contain the molecules in the simulation. It then constructs a series of tan counting boxes along the length to count the molecules. It also constructs a series of light blue counting planes to measure the flux. Molecules are released at the center of the box at t=0 and diffuse during the simulation.

14.1.4.5.1. Fick’s Law Construction Script¶
The following script builds the CellBlender model from scratch.
# Import cellblender to get the data model and project directory functions
import cellblender as cb
# Get a copy of the data model and change directories to the project directory for any file I/O
dm = cb.get_data_model(geometry=True)
old_location = cb.cd_to_project()
# Print some information about the data model
print ( "###############################################################" )
print ( "Data Model Top Level Keys = " + str(dm.keys()) )
print ( "MCell Keys = " + str(dm['mcell'].keys()) )
print ( "###############################################################" )
# Define parameters and set default values (these will also be available as local variables in this script)
# The format for each parameter is: Name, Value, Units, Description
# All values are given as strings!!!
pars = [
# These control the geometry of the model
['n', "40", '', 'Number of segments to sample along the x dimension'],
['lx', "2.0", 'um', 'Length x = Total length of sample box in x dimension'],
['ly', "0.2", 'um', 'Length y = Total length of sample box in y dimension'],
['lz', "0.2", 'um', 'Length z = Total length of sample box in z dimension'],
['ext', "0.02", 'um', 'Extended length for counting boxes and counting planes'],
['tol', "0.995", 'um', 'Scale factor of sampling boxes to avoid coincident faces (0.995 works well)'],
['rtol', "0.001", 'um', 'Release plane tolerance - smaller is closer to ideal (0.001 works well)'],
# These control the instrumentation of the model
['plot_segment_counts', "1", '', 'Plot count of vm molecules in each segment when non-zero'],
['plot_front_crossings', "0", '', 'Plot front crossings of vm molecules when non-zero'],
# These describe the behavior of the model
['dc', "5e-6", 'cm^2/sec', 'Diffusion Constant of Molecules'],
['nrel', "1000", 'Count', 'Number of molecules to release'],
# These control the simulation itself
['iters', "600", '', 'Number of iterations to run'],
['seeds', "10", '', 'Number of seeds to run'],
['dt', "1e-6", 'sec', 'Time step for each iteration of the simulation'],
]
# Update local parameter list values from the existing data model with user-modified settings BEFORE regenerating it
dm_par_list = dm['mcell']['parameter_system']['model_parameters']
for dm_par in dm_par_list:
print ( "Data Model Parameter " + dm_par['par_name'] + " = " + dm_par['par_expression'] )
for p in pars:
if dm_par['par_name'] == p[0]:
# Update the local expression based on the parameter found in the incoming data model
p[1] = dm_par['par_expression']
p[2] = dm_par['par_units']
p[3] = dm_par['par_description']
# Create the local variables from the updated values to use in this script
for p in pars:
locals()[p[0]] = eval(p[1])
# Create the new mcell data model inside the existing data model (this deletes the previous mcell data model)
dm['mcell'] = { 'data_model_version' : "DM_2014_10_24_1638" }
# Restore the parameters that were either initialized from scratch or preserved from the previous data model
dm['mcell']['parameter_system'] = { 'model_parameters':[] } # Parameters are currently unversioned
for p in pars:
dm['mcell']['parameter_system']['model_parameters'].append ( { 'par_name':p[0], 'par_expression':p[1], 'par_units':p[2], 'par_description':p[3] } )
# Define a function to make either a plane or a box from its center and lengths in each dimension (one zero dimension gives a plane)
def make_obj ( center_x, center_y, center_z, len_x, len_y, len_z ):
obj = {}
obj['vertex_list'] = []
obj['element_connections'] = []
if len_x == 0:
# Make a plane perpendicular to the x axis
obj['vertex_list'].append ( [ center_x, center_y-(len_y/2.0), center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x, center_y-(len_y/2.0), center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x, center_y+(len_y/2.0), center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x, center_y+(len_y/2.0), center_z-(len_z/2.0) ] )
obj['element_connections'].append ( [ 0, 2, 1 ] )
obj['element_connections'].append ( [ 0, 3, 2 ] )
elif len_y == 0:
# Make a plane perpendicular to the y axis
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y, center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y, center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y, center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y, center_z-(len_z/2.0) ] )
obj['element_connections'].append ( [ 0, 2, 1 ] )
obj['element_connections'].append ( [ 0, 3, 2 ] )
elif len_z == 0:
# Make a plane perpendicular to the z axis
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y-(len_y/2.0), center_z ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y+(len_y/2.0), center_z ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y+(len_y/2.0), center_z ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y-(len_y/2.0), center_z ] )
obj['element_connections'].append ( [ 0, 2, 1 ] )
obj['element_connections'].append ( [ 0, 3, 2 ] )
else:
# Make a box
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y+(len_y/2.0), center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y-(len_y/2.0), center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y-(len_y/2.0), center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y+(len_y/2.0), center_z-(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y+(len_y/2.0), center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x+(len_x/2.0), center_y-(len_y/2.0), center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y-(len_y/2.0), center_z+(len_z/2.0) ] )
obj['vertex_list'].append ( [ center_x-(len_x/2.0), center_y+(len_y/2.0), center_z+(len_z/2.0) ] )
obj['element_connections'].append ( [ 1, 2, 3 ] )
obj['element_connections'].append ( [ 7, 6, 5 ] )
obj['element_connections'].append ( [ 4, 5, 1 ] ) # Right end
obj['element_connections'].append ( [ 5, 6, 2 ] )
obj['element_connections'].append ( [ 2, 6, 7 ] ) # Left end
obj['element_connections'].append ( [ 0, 3, 7 ] )
obj['element_connections'].append ( [ 0, 1, 3 ] )
obj['element_connections'].append ( [ 4, 7, 5 ] )
obj['element_connections'].append ( [ 0, 4, 1 ] ) # Right end
obj['element_connections'].append ( [ 1, 5, 2 ] )
obj['element_connections'].append ( [ 3, 2, 7 ] ) # Left end
obj['element_connections'].append ( [ 4, 0, 7 ] )
return obj
# Add materials for the objects
dm['mcell']['materials'] = { 'material_dict' : {} } # Materials are currently unversioned
dm['mcell']['materials']['material_dict']['box_color'] = { 'diffuse_color' : {'a':0.3, 'r':0.2, 'g':0.4, 'b':1.0} }
dm['mcell']['materials']['material_dict']['rel_color'] = { 'diffuse_color' : {'a':0.2, 'r':0.9, 'g':0.7, 'b':0.5} }
dm['mcell']['materials']['material_dict']['vol_color'] = { 'diffuse_color' : {'a':0.1, 'r':0.9, 'g':0.7, 'b':0.5} }
dm['mcell']['materials']['material_dict']['plane_color'] = { 'diffuse_color' : {'a':0.7, 'r':0.5, 'g':0.7, 'b':1.0} }
# Create container objects for geometrical objects and model objects
dm['mcell']['geometrical_objects'] = {} # Geometrical objects are currently unversioned
dm['mcell']['model_objects'] = { 'data_model_version':"DM_2014_10_24_1638" }
# Each container also includes a list
dm['mcell']['geometrical_objects']['object_list'] = []
dm['mcell']['model_objects']['model_object_list'] = []
# Add objects to the lists
# Make the main box for diffusing the molecules
box = make_obj ( 0, 0, 0, 10*lx, ly, lz ) # Make the box much longer to reduce boundary effects from absorptive ends
box['name'] = 'box'
box['material_names'] = [ 'box_color' ]
# Make the thin box for releasing the molecules
rel = make_obj ( 0, 0, 0, rtol, ly-rtol, lz-rtol )
rel['name'] = 'rel'
rel['material_names'] = [ 'rel_color' ]
# Make the surface regions for the two absorptive ends to keep them from accumulating
box['define_surface_regions'] = []
box['define_surface_regions'].append ( { 'name':"left_end", 'include_elements':[ 4, 10 ] } )
box['define_surface_regions'].append ( { 'name':"right_end", 'include_elements':[ 2, 8 ] } )
# Add the box to the geometrical objects and the model objects
dm['mcell']['geometrical_objects']['object_list'].append ( box )
dm['mcell']['model_objects']['model_object_list'].append ( { 'name':box['name'] } )
dm['mcell']['geometrical_objects']['object_list'].append ( rel )
dm['mcell']['model_objects']['model_object_list'].append ( { 'name':rel['name'] } )
# Make the counting boxes and planes as requested by the parameter flags
for i in range(n):
x = (i - ((n-1)/2.0)) / (n/lx)
if plot_segment_counts != 0:
box = make_obj ( x, 0, 0, tol*(lx/n), ly+ext, lz+ext )
box['name'] = 'vol_%03d' % i
box['material_names'] = [ 'vol_color' ]
dm['mcell']['geometrical_objects']['object_list'].append ( box )
dm['mcell']['model_objects']['model_object_list'].append ( { 'name':box['name'] } )
if (plot_front_crossings != 0) and (i > 0):
plane = make_obj ( x-(lx/(2*n)), 0, 0, 0.0, ly+ext+ext, lz+ext+ext )
plane['name'] = 'plane_%03d' % i
plane['material_names'] = [ 'plane_color' ]
dm['mcell']['geometrical_objects']['object_list'].append ( plane )
dm['mcell']['model_objects']['model_object_list'].append ( { 'name':plane['name'] } )
# Create a molecule list and create a "vm" molecule along with its display properties in that list
dm['mcell']['define_molecules'] = { 'data_model_version' : "DM_2014_10_24_1638" }
mol = { 'mol_name':"vm", 'mol_type':"3D", 'diffusion_constant':"dc", 'data_model_version':"DM_2016_01_13_1930" }
mol['display'] = {'color':[0.0,1.0,0.0], 'emit':1.0, 'glyph':"Cube", 'scale':0.5 }
dm['mcell']['define_molecules']['molecule_list'] = [ mol ]
# Create a release site
rel_site = {
'name' : "center_rel",
'molecule' : "vm",
'quantity' : "nrel",
'quantity_type' : "NUMBER_TO_RELEASE",
'release_probability' : "1",
'shape' : "OBJECT",
'object_expr' : "rel",
'orient' : ";",
'pattern' : "",
'location_x' : "0",
'location_y' : "0",
'location_z' : "0",
'site_diameter' : "0",
'stddev' : "0",
'data_model_version' : "DM_2015_11_11_1717"
}
dm['mcell']['release_sites'] = { 'release_site_list':[ rel_site ], 'data_model_version':"DM_2014_10_24_1638" }
# Define surface classes
dm['mcell']['define_surface_classes'] = { 'surface_class_list':[], 'data_model_version':"DM_2014_10_24_1638" }
# Use a table to construct the various classes with associated properties
surf_classes = [
[ 'transp', 'vm_transp', ';', "TRANSPARENT", "0" ],
[ 'absorb', 'vm_absorb', ';', "ABSORPTIVE", "0" ] ]
# Loop through the table and add each class to the data model
for c in surf_classes:
sc_prop = { 'data_model_version':"DM_2015_11_08_1756",
'name':c[1],
'affected_mols':"SINGLE",
'molecule':"vm",
'surf_class_orient':c[2],
'surf_class_type':c[3],
'clamp_value':c[4]
}
sc_entry = { 'data_model_version':"DM_2014_10_24_1638",
'name':c[0],
'surface_class_prop_list':[ sc_prop ]
}
dm['mcell']['define_surface_classes']['surface_class_list'].append ( sc_entry )
# Assign the surface classes with the "modify_surface_regions" key
dm['mcell']['modify_surface_regions'] = { 'modify_surface_regions_list':[], 'data_model_version': "DM_2014_10_24_1638" }
# Modify the left end to be absorptive
dm['mcell']['modify_surface_regions']['modify_surface_regions_list'].append (
{
'name':"absorb left",
'object_name':"box",
'region_name':"left_end",
'surf_class_name':"absorb",
'region_selection':"SEL",
'data_model_version':"DM_2015_11_06_1732"
} )
# Modify the right end to be absorptive
dm['mcell']['modify_surface_regions']['modify_surface_regions_list'].append (
{
'name':"absorb right",
'object_name':"box",
'region_name':"right_end",
'surf_class_name':"absorb",
'region_selection':"SEL",
'data_model_version':"DM_2015_11_06_1732" } )
# Modify the release box, all counting boxes, and counting planes (if any) to be transparent
dm['mcell']['modify_surface_regions']['modify_surface_regions_list'].append (
{
'name':"transp rel",
'object_name':"rel",
'region_name':"",
'surf_class_name':"transp",
'region_selection':"ALL",
'data_model_version':"DM_2015_11_06_1732"
} )
for i in range(n):
if plot_segment_counts != 0:
name = 'vol_%03d' % i
dm['mcell']['modify_surface_regions']['modify_surface_regions_list'].append (
{
'name':"transp "+name,
'object_name':name,
'region_name':"",
'surf_class_name':"transp",
'region_selection':"ALL",
'data_model_version':"DM_2015_11_06_1732"
} )
if (plot_front_crossings != 0) and (i > 0):
name = 'plane_%03d' % i
dm['mcell']['modify_surface_regions']['modify_surface_regions_list'].append (
{
'name':"transp "+name,
'object_name':name,
'region_name':"",
'surf_class_name':"transp",
'region_selection':"ALL",
'data_model_version':"DM_2015_11_06_1732"
} )
# Define the counting output
dm['mcell']['reaction_data_output'] = {
'data_model_version':"DM_2014_10_24_1638",
'reaction_output_list':[],
'rxn_step':"10*dt",
'combine_seeds':False,
'mol_colors':True,
'plot_layout':" plot ",
'plot_legend':"x",
'mol_colors':False
}
dm['mcell']['reaction_data_output']['reaction_output_list'].append (
{
'data_model_version':"DM_2015_10_07_1500",
'name':"vm in box",
'rxn_or_mol':"Molecule",
'mdl_string':"",
'mdl_file_prefix':"",
'count_location':"Object",
'object_name':"box",
'region_name':"",
'reaction_name':"",
'molecule_name':"vm"
} )
# Create the counting structures for the counting object as requested
for i in range(n):
if plot_segment_counts != 0:
name = 'vol_%03d' % i
if plot_segment_counts != 0:
dm['mcell']['reaction_data_output']['reaction_output_list'].append (
{
'data_model_version':"DM_2015_10_07_1500",
'name':"vm in "+name,
'rxn_or_mol':"Molecule",
'mdl_string':"",
'mdl_file_prefix':"",
'count_location':"Object",
'object_name':name,
'region_name':"",
'reaction_name':"",
'molecule_name':"vm"
} )
if (plot_front_crossings != 0) and (i > 0):
name = 'plane_%03d' % i
mdl_string = "COUNT[vm,Scene."+name+",FRONT_CROSSINGS]"
dm['mcell']['reaction_data_output']['reaction_output_list'].append (
{
'data_model_version':"DM_2015_10_07_1500",
'name':"MDL: "+mdl_string,
'rxn_or_mol':"MDLString",
'mdl_file_prefix':name+"_front_cross",
'mdl_string':mdl_string,
'count_location':"World",
'object_name':"",
'region_name':"",
'reaction_name':"",
'molecule_name':""
} )
# Set up the simulation running parameters
dm['mcell']['initialization'] = { 'data_model_version':"DM_2014_10_24_1638" }
dm['mcell']['initialization']['iterations'] = "iters"
dm['mcell']['initialization']['time_step'] = "dt"
dm['mcell']['simulation_control'] = { 'data_model_version': 'DM_2016_04_15_1430' }
dm['mcell']['simulation_control']['start_seed'] = '1'
dm['mcell']['simulation_control']['end_seed'] = 'seeds'
# Return to the previous directory and replace the existing data model with this modified version
cb.cd_to_location ( old_location )
cb.replace_data_model ( dm, geometry=True )
14.1.4.5.2. Fick’s Law Plotting Script¶
The following plot series (animation) compares the average of 50 MCell runs (Start Seed=1, End Seed=50, 10,000 molecules released) at various points in time to plots of the expected theoretical results at those same points in time.
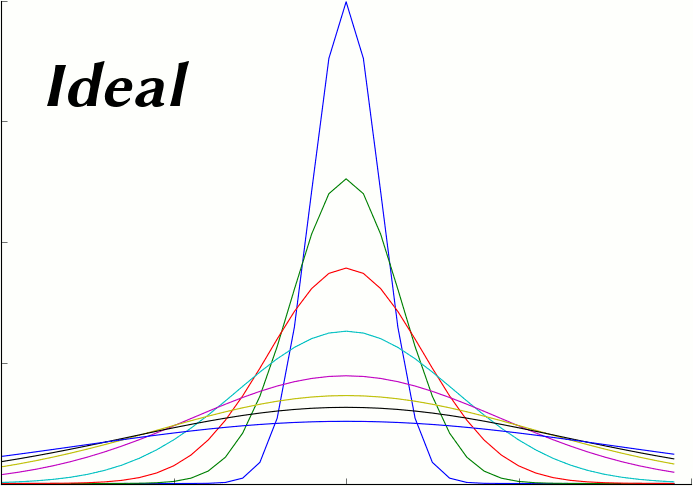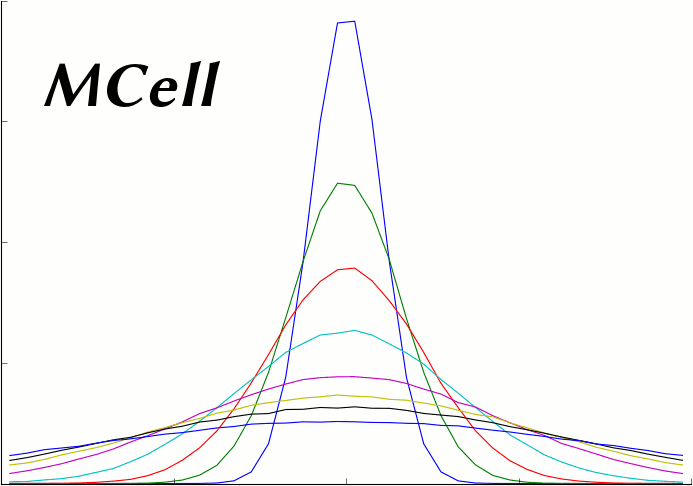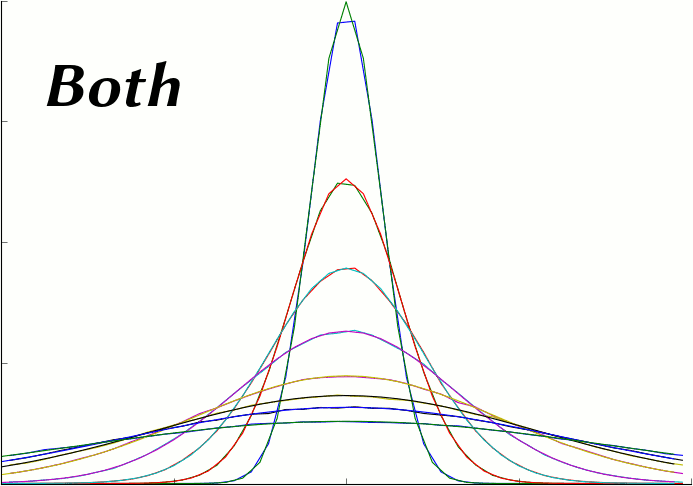
This script generates the data needed for that plot. It reads and averages the data from the MCell runs and generates a series of plot files from that output. It also generates a series of plot files from the expected analytic solution with the same parameters as the MCell runs. Because this script uses the same parameters used to generate the simulation, it will produce the correct plot even when the simulation parameters are varied.
import math
import os
from numpy import fromfile
import cellblender as cb
dm = cb.get_data_model()
old_location = cb.cd_to_project()
pars = [
# These control the geometry of the model
['n', "40", '', 'Number of segments to sample along the x dimension'],
['lx', "2.0", 'um', 'Length x = Total length of sample box in x dimension'],
['ly', "0.2", 'um', 'Length y = Total length of sample box in y dimension'],
['lz', "0.2", 'um', 'Length z = Total length of sample box in z dimension'],
['ext', "0.02", 'um', 'Extended length for counting boxes and counting planes'],
['tol', "0.995", 'um', 'Scale factor of sampling boxes to avoid coincident faces (0.995 works well)'],
['rtol', "0.001", 'um', 'Release plane tolerance - smaller is closer to ideal (0.001 works well)'],
# These control the instrumentation of the model
['plot_segment_counts', "1", '', 'Plot count of vm molecules in each segment when non-zero'],
['plot_front_crossings', "0", '', 'Plot front crossings of vm molecules when non-zero'],
# These describe the behavior of the model
['dc', "5e-6", 'cm^2/sec', 'Diffusion Constant of Molecules'],
['nrel', "1000", 'Count', 'Number of molecules to release'],
# These control the simulation itself
['iters', "500", '', 'Number of iterations to run'],
['seeds', "10", '', 'Number of seeds to run'],
['dt', "1e-6", 'sec', 'Time step for each iteration of the simulation'],
]
# Update local parameter list values from the existing data model with user-modified settings BEFORE generating plot data
dm_par_list = dm['mcell']['parameter_system']['model_parameters']
for dm_par in dm_par_list:
print ( "Data Model Parameter " + dm_par['par_name'] + " = " + dm_par['par_expression'] )
for p in pars:
if dm_par['par_name'] == p[0]:
# Update the local expression based on the parameter found in the incoming data model
p[1] = dm_par['par_expression']
p[2] = dm_par['par_units']
p[3] = dm_par['par_description']
# Create the local variables from the updated values to use in this script
for p in pars:
locals()[p[0]] = eval(p[1])
plot_iters = [ 10, 25, 50, 100, 200, 300, 400, 600 ]
start_seed = 1
end_seed = seeds
start_vol = 0
end_vol = n - 1
num_vols = 1 + end_vol - start_vol
# Note: Neither 'iterations' nor 'time_step' can use CellBlender parameters!!
iters = eval(dm['mcell']['initialization']['iterations'])
dt = eval(dm['mcell']['initialization']['time_step'])
react_files_dir = "mcell" + os.sep + "react_data"
react_files_seeds = os.listdir(react_files_dir)
print ( "files: " + str(react_files_seeds) )
parent_dir = os.getcwd().split(os.sep)[-1]
# Remove all plots from the data model so they're not cumulative when this script is re-run
dm['mcell']['reaction_data_output']['reaction_output_list'] = [] # Comment this line to NOT remove previous plots
def make_file_plot ( file_name ):
blender_relative_name = "//" + parent_dir + os.sep + fname
reaction_out = { 'data_model_version':"DM_2016_03_15_1800" }
reaction_out['rxn_or_mol'] = "File"
reaction_out['molecule_name'] = ""
reaction_out['reaction_name'] = ""
reaction_out['object_name'] = ""
reaction_out['region_name'] = ""
reaction_out['mdl_file_prefix'] = ""
reaction_out['mdl_string'] = ""
reaction_out['count_location'] = "World"
reaction_out['plotting_enabled'] = True
reaction_out['data_file_name'] = blender_relative_name
reaction_out['name'] = "FILE:" + blender_relative_name
return reaction_out
area = ly * lz
# Generate the MCell plots
for plot_iter in plot_iters:
print ( "Generating concentration curve for " + str(plot_iter) )
fname = "concentration_%d.txt" % plot_iter
f = open ( fname, "w" )
dm['mcell']['reaction_data_output']['reaction_output_list'].append ( make_file_plot(fname) )
points = []
for vol in range(start_vol,1+end_vol):
count = 0.0
sx0 = (1+end_vol-start_vol)/2
sx = (vol - sx0)
vx = sx * lx / n
for seed in range(start_seed,1+end_seed):
file_name = react_files_dir + os.sep + ("seed_%05d" % seed) + os.sep + ("vm.vol_%03d.dat" % vol)
data = fromfile ( file_name, sep=' ' )
x = data[0::2]
y = data[1::2]
count = count + y[plot_iter]
averaged_count = count/(1+end_seed-start_seed)
conc = averaged_count / (tol*lx/n) # This should be molecules per micron (length)
f.write ( str(vx+(lx/(2.0*n))) + " " + str(conc) + "\n" )
f.close()
# Generate the analytic plots
for plot_iter in plot_iters:
print ( "Generating analytic curve for " + str(plot_iter) )
fname = "concentration_ideal_%d.txt" % plot_iter
f = open ( fname, "w" )
dm['mcell']['reaction_data_output']['reaction_output_list'].append ( make_file_plot(fname) )
t = plot_iter * dt
for vol in range(start_vol,1+end_vol):
sx0 = (1+end_vol-start_vol)/2
sx = (vol - sx0)
x = sx * lx / n
N = nrel * math.exp(-(x*x/(4*dc*1e8*t))) / (2 * math.sqrt(math.pi*dc*1e8*t))
f.write ( str(x) + " " + str(N) + "\n" )
f.close()
# Configure the plotter for one page / one plot (uses a single space):
dm['mcell']['reaction_data_output']['plot_layout'] = ' '
cb.cd_to_location ( old_location )
cb.replace_data_model ( dm )